Tools & Libraries
On this page we present libraries and tools working with SED-ML. If you would like your software tool to be listed here please fill out the survey.
SED-ML libraries
| Name | Languages | SED-ML Versions | Description | Screenshot |
|---|---|---|---|---|
| BioSimulators utils | Python |
|
High-level library for reading, writing, validating and executing SED-ML documents, including documents inside COMBINE/OMEX archives. Also provides a command-line application for validating and executing SED-ML documents inside COMBINE/OMEX archives with BioSimulators-compliant simulation tool. | |
| clibsedml | C |
|
Portable C library for SED-ML. | 
|
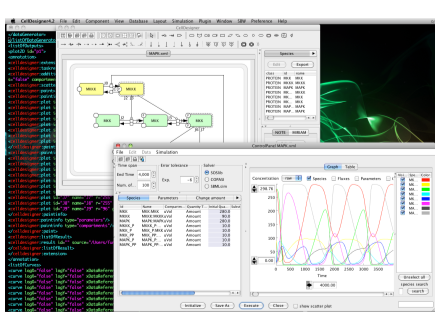
| jlibsedml | Java |
|
An extensible open source Java library to create, manipulate, validate and store SED-ML files. jlibsedml also provides XPath support and an execution framework for running simulations and post-processing the results - clients provide the simulation implementation. | 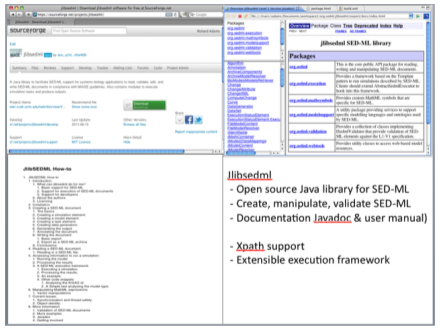
|
| libSEDML | C++, C# (Swig), Java (Swig), Perl (Swig), Python (Swig), R (Swig), Ruby (Swig) |
|
C++ library for reading and writing SED-ML. Swig languange bindings for C#, Java, Python, R and Ruby. | |
| libSedML (old version) | .NET library |
|
libSedML is a set of .NET libraries for supporting SED-ML. The core library libSedML supports reading, validating and writing of SED-ML descriptions, along with all necessary utility functions for resolving models and XPath expressions. Two additional libraries are included: libSedMLRunner, which allows to schedule and execute simulation experiments encoded in SED-ML files using either RoadRunner# (Bergmann et al., 2006) or a variety of simulators exposed through the Systems Biology Workbench (SBW, Bergmann et al., 2006a) like iBioSim (Myers et al., 2009) and COPASI (Hoops et al., 2006). A third library, libSedMLScript, provides a script based language for defining SED-ML experiments. | 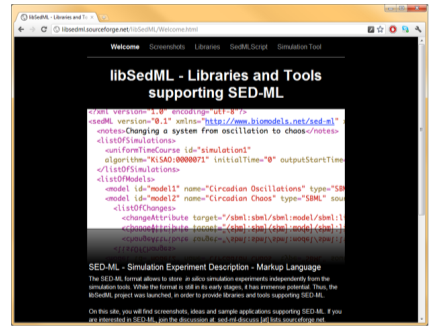
|
| phraSED-ML | C++, Python (Swig) |
|
A paraphrased, human-readable adaptation of SED-ML, phraSED-ML language allows modelers to use simple text commands to encode various elements of SED-ML (models, tasks, simulations, and results) in a format easy to read and modify. The library can translate this script to SED-ML for use in other softwares. |
SED-ML tools
| Name | Model Languages | SED-ML Versions | Description | Screenshot |
|---|---|---|---|---|
| BioSimulators |
|
|
Registry of simulation tools with information about the model formats, simulation formats, modeling frameworks, and simulation algorithms that they support. Many simulation tools are available as Docker images with standardized entrypoints for executing SED-ML files in COMBINE/OMEX archives. | |
| BioSimulators test suite |
|
|
Tool for checking that simulation tools execute SED-ML files and COMBINE archives consistently with the specifications of these formats. | |
| BioSimulators utils |
|
|
Command-line application for validating and executing SED-ML documents inside COMBINE/OMEX archives with BioSimulators-compliant simulation tool. Also provides a high-level Python library for reading, writing, validating and executing SED-ML documents, including documents inside COMBINE/OMEX archives. | |
| COPASI |
|
|
Software application for simulation and analysis of biochemical networks and their dynamics. COPASI is a stand-alone program that supports models in the SBML standard and can simulate their behavior using ODEs or Gillespie`s stochastic simulation algorithm; arbitrary discrete events can be included in such simulations. COPASI supports impor and export of SED-ML files. | |
| Copasi2SedML |
|
|
Library and tools that allow to convert a COPASI file containing a time course task to SED-ML L1V1. This work is based on the COPASI C# bindings and libSedML.. | 
|
| iBioSim |
|
|
Tool for the modeling, analysis, and design of genetic circuits. iBioSim support models representing metabolic networks, cell-signaling pathways, and other biological and chemical systems. iBioSim supports importing and exporting models specified using the Systems Biology Markup Language (SBML). It can import all levels and versions of SBML and is able to export Level 3 Version 1. Simulations of SBML models based on SED-ML is supported. | |
| JWS Online |
|
|
Tool for the construction, modification and simulation of kinetic models and for the storage of curated models. It provides a selection of simulation experiments as SED-ML and COMBINE archives. | 
|
| OpenCOR |
|
|
Open source cross-platform modelling environment. OpenCOR is a replacement for COR. | 
|
| runBioSimulations |
|
|
Web application and REST API for executing SED-ML documents in COMBINE/OMEX archives and visualizing their results. runBioSimulations can execute simulations using all of the standardized simulation tools in the BioSimulators registry. Together, these tools support simulations for multiple model formats, modeling frameworks, and over 40 simulation algorithms. | |
| SBML Testsuite |
|
|
Conformance testing system for SBML. It allows developers and users to test the degree and correctness of SBML support provided in an SBML-compatible software package. All tests are available also as SED-ML files. | 
|
| sbmlsim |
|
|
Collection of Python utilities to simulate to simplify simulations with SBML models implemented on top of roadrunner. Features include among others simulation experiments, simulation reports parameter fitting. | |
| Systems Biology Simulation Core Library (SBSCL) |
|
|
Efficient and exhaustive Java library of methods to interpret the content of models encoded in the Systems Biology Markup Language (SBML) and its numerical solution. SBSCL is based on JSBML. SBSCL can also read SED-ML files. | 
|
| SBW Simulation Tool |
|
|
Extensible, interactive environment for carrying out simulation experiments of SBML models. This project can use RoadRunner or SBW enabled simulators to carry out simulations.The simulation tool allows you to execute the simulations directly, with support for remote models (even in BioModels.net) and the SED-ML archive. | 
|
| SED-ED |
|
|
Visual editor for viewing, editing and validating SED-ML documents using a graphical workflow notation. SED-ED previews model changes for any XML based model, generates XPath expressions for targetting model elements, displays and edits notes in an embedded browser, works with SEDX archives as well as plain SEDML files, retrieves models from Biomodels using Miriam URNs, provides standard undo/redo/copy/paste/align functionality for editing. SED-ED is available either as a standalone application, an Eclipse plugin (update site http://www.sbsi.ed.ac.uk/update) or an SBSIVisual plugin. | 
|
| SED-ML Script Editor |
|
|
Allows to construct SED-ML documents using a simplified script language. Starting out by directly importing SBML or CellML models an initial script is generated that can be simulated. From there it is easy to modify it to suite your needs. | 
|
| SED-ML Web Tools |
|
|
Online application providing support to edit, simulate and validate SED-ML files online. | 
|
| tellurium |
|
|
Python environment based on spyder2 IDE. It combines a number of existing libraries, including libSBML, libRoadRunner (including libStruct), libAntimony, and is extensible via telPlugins. | 
|
| VCell |
|
|
Comprehensive platform for modeling cell biological systems that is built on a central database and disseminated as a web application. |
© 2017-2021 sed-ml.org. Report website issue